Enzymes
- Created by: rosieevie
- Created on: 13-01-17 18:44
Enzyme Kinetics
Study of enzymes to characterise the rates/steps of catalysis to compare them
Must collect experimentsal data - change in concentration of sub/react over time
Can use a spectrophotometer which measures absorbance change of radiation e.g. visible/UV
Reaction Rate
In an equilibrium reaction - the reaction appears to halt as conc. remains constant. Rate = Zero
Reaction rate depends on speed of reaction and con. of reactants/substrates
Each reaction has a rate constant (k)
FORWARD: (Ks->p) x [S]
BACKWARD (Kp->s) x [P]
Whichever number is higher determines the reaction direction (num. equal at equilibrium)
Enzymes change k by decreasing activation energy (provide an alternative route w/ less energy = more molecules with sufficient energy = more reactions)
Reactions can be 'pushed' in diff. directions by changing concs. Equilibrium ratio will stay the same
Measuring Reaction Rates
In a reaction [P] can increase over time but not constant rate.
Therefore take zero value and extend = reaction velocity (v mol/min or mol/s)
Also measure enzyme activity (umol/min) or specific activity (umol/min/mg)
Enzyme activity/Total amount of protein = indication of purity
Enzyme-catalysed reactions have two steps:
E + S --binding--> ES ---catalysis--> E + P
Steps may occur at different speeds
Michaelis-Menten Model 1
K+2 is called Kcat and K-2 is ignored
K-1/K+1 = [E][S]/[ES] = Dissociation constant (Kd) = Enzymes affinity for [ES]
Small Kd = high affinity [ES]
Steady-State Kinetics
Assumptions:
- [ES] constant because reactions occur so quickly
- [S]>>[E] so [S] is constant - no overall decrease in sub. concentration so rate not effected
Michaelis-Menten Curve
Vmax - the maximum velocity/rate which an enzyme catalyses a reaction (when all enzymes saturated)
- Helps to work out Km
- Km = Vmax/2
- The line will never reach Vmaz as the sub. would no longer dissolve in solution first
Types of Reaction
Anabolic/synthetic - create something body needs
Catabolic/degradative - break molecules
Interconverstion - reveresible reaction where same enzyme is used. Substrate and product in equilibrium - balance
Coenzymes
Organic molcules which provide/remove groups e.g. H+ ions.
Co-substrates as can bind to active site
Examples - NADH->NAD+
Enzyme Classification
- Most names end in -ase
- Some have common names e.g. trypsin
- Named for reaction or substrate:
- Kinase - transfer (PO4)3- from ATP to OH groups
- Phosphatase - remove (PO4)3- to leave OH (opposite of kinase)
- Dehydrogenases - remove H with NAD+
- Glycogen synthase - makes glycogen
- Every enzyme has unique enzyme commission number (4 digits)
- 6 enzyme classes (refer to reaction):
- Oxidoreductases - transfer electrons (H/H-)
- Transferases - transfer chemical groups
- Hydrolases - break bonds with water
- Lyases - reactions involve double bonds
- Isomerases - transfer groups within a molecule (rearranges bonds)
- Ligases - formation of bonds using ATP
Lysosomes
Destroy and recycle cells
Acid sensitive, require low pH - protects rest of cell from digestion
Acid phosphate marker enzyme for lysosomes (tell you are looking at a lysosome)
Advantages
Maintain pace of life and maintain required conditions for life e.g. pH, body temp
- Reusable - save resources
- Specific
- Efficient - 100% yield
- Controllable - start/inhibit reactions
The Active Site
Small part of enzyme - rest just holds active site in place
In 3D arrangement the active site is scattered
Contains binding and catalytic residues - source of substrate and reaction specificity
Small part of substrate enters active site - sometimes means not so specific e.g. medicines
Reaction Specificity
Determined by:
- 3D residue arrangement - close enough to right bond
- Chemical properties of residues e.g. positive group only does certain things
Stereospecificity - Optical Isomers
- Catalytic triad - only 3 amino acids perform catalysis
- Active site has 3 recognition points
- If there are stereoisomers only one form of the amino acid will be recognised
Active site also can contain:
- Metal cofactors e.g. Mg2+, Zn2+
- Prosthetic groups - organic groups required for overall function e.g. haem
Substrate Specificity
Substrate specificity is affected by siz/shape filler and binding affinity
Lock and Key (Fischer)
- Shape of active site complementary to substrate
- If not, no enzyme-substrate complex
Induced Fit (Koschland)
- Mututal conformational change of substrate and enzyme
- Pull together as substrate enters active site due to bonds
- Due to any type of bond e.g. ionic, van der Waals, hydrophobic
What are Enzymes?
Enzyme - one or more polypeptide chain forming a catalytic active site
Substrate - molecule which binds to the active site and undergoes a chemical reaction
Product - result of enzyme action
Enzyme roles:
- Digestion (pepsin)
- Blood clotting (thrombin)
- Control blood pressure (ACE)
- Defence (lysosyme)
- Breakdown of toxins (cytochrome)
- Routine cell processes
Lineweaver-Burk Plot
Used to calculate Vmax using recipricols - turns plots into a straight line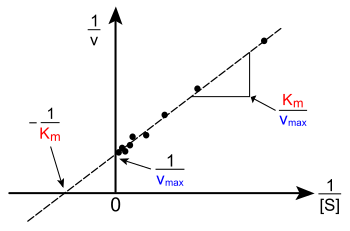
Vmax - the maximum possible rate (when all enzymes have active sites filled)
Vmaz = Kcat x [E] ---> Kcat = Vmax/[E] use Vmax to determine Kcat
Km
Km:
- [S] which gives half maximum rate
- [S] at which half of the enzymes have formed enzyme substrate complexes (allows max. flexibility to change reaction rate)
A low Km suggests a high affinity for the substrate
Biological Significances
Vmax
- Not signifiicate - need an infinate sub. conc. to retain this
- Usually low - saturating [S] is unusual
- Except when drinking excessively
Km
- Usually high - [S] in cell often close to Km
- Each substrate has a different Km for the same enzyme
- Methanol posioning - treated with ethanol as enzyme has a high affinity for it
- Determines how active an enzyme is at a particular concentration
Comparing Enzymes
- Turnover number - catalytic rate constant (Kcat) - number of reactions per second
- Enzyme efficiency - catalytic speed if Kcat/Km > 1x108 V is limited by diffusion of substrate not enzyme itself (kinetic perfection)
- Enzyme potency - how many times faster reaction is with enzyme
Sigmoidal Curves
- Suggests a multiple subunit enzyme with co-operative changes in substrate affinity between subunits
- Substrate binding to one site increases affinity at another
- Concentration directly regulates enzyme activity
- Advantage because controllable
Controlling Enzyme Activity
- Changing temperature on V - Increase increases V up until denaturing
- Changing pH on V - Indi. enzymes have optimum pH
- Slight drop - enzyme losing positive/negative charges
- Big drop - denaturing
- Changing [E] on V - switching genes on and off changes concentration
- How you get over enzyme saturation (only applies to a fixed conc.)

- How you get over enzyme saturation (only applies to a fixed conc.)
Et = [E]
Direct Enzyme Regulation
Covalent
Most digestive enzymes synthesised in inactive form (damaging) - activated by peptide chain cleavage (irreversible reaction) = ZYMOGENS
Phosphorylisation (reversible) - addition of phosphate group by kinase which distorts active site, denaturing it
Non-Covalent - Allosteric Enzymes
Reversible binding of molecules to specific sites NOT active site -> increase/decrease activity
- K-type enzyme - effects binding and changes Km
- Extra polypeptide chain where regulatory molecule reversibly binds
- Allosteric activator - higher conc leads to increased activity
- Allosteric inhibitor - higher conc leads to decreased activity
- V-type regulation - effects catalysis and changes Vmax
- Change ability of enzyme to catalyse reaction
Competitive Inhibitors
Enzyme inhibitors reduce enzyme activity - either reversible or irreversible
- Prevents entry of substrate
- Binds in active site or away from active site
- Lowers V
- Vmax same (increase [S] = less effect of inhibitor) - delayed reaction though
- Km larger = weaker affinity
- Effect similar to less substrate due to competition
- V = Vmax x [S]/((Km x If) + [S])
Non-Competitive Inhibitors
- Reaction can never occur even though substrate can bind
- Binds away from active site
- Lowers V
- Smaller Vmax - never reach same
- Same Km - binding affinity unchanged
- Same effect as enzyme conc. decreased
- V = ((Vmax/If) x [S])/Km + [S]
Uncompetitive Inhibitors
- Bind once substrate is in active site
- Locks substrate in active site but no reaction
- Vmax is smaller
- Km is smaller
- V = Vmax x [S]/(Km + ([S] x If))
Using Inhibitors as Lab Tools
- Enzyme structure - inhibitors can stabalise structure and crystalise 'active' form
- Enzyme purification - coat affinity column beads with inhibitors = only enzyme sticks
- Active site investigation
- Pseudosubstrates bind and irreversibly alter active site
- Protective masks - determine where active site is
Enzyme Binding Methods
Difficult to explain kinetics with multiple substrate and product reactions - sometimes order needed or random
Sequential Method - both have to bind before reaction occurs
E + A + B --> EAB ---> ECD ---> E + C + D
- Order - smaller substrate may have to bind first
- Random - either can bind first
Ping-Pong Method - substrates never together in the active site
E + A --> EA ---> EC --C leaves --> E' ---> E'B ----> E'D ---> E + D
Covalent bonds to E formed during process
Transition state must form before reaction occurs - can take time. Binding steps partially offset activation energy
Enzyme Strategies to Increase Reaction Rate
General Strategies
- Position reactions into correct orientation for interaction
- Distort reactans making bonds less stable
- Stabilises transition state - prefered binding (stronger interactions)
- Chnages environment to favour reaction e.g. pH, hydrophobic, salinity
Best inhibitor drugs resemble transition state
Specific Chemical Strategies
- Covalent catalysis - active site residue reacts with substrate
- Acid-base catalysis - active site residues accept/donate H+ ions
- Metal ion catalysis - concentrated positive charges
Combo of the 2 which lowers overall reaction energy
EXAMPLE - PROTEOLYSIS (protein breakdown)
Proteases break down stable peptide bonds efficiently
4 main types focused (all have different substrates and different specificity pockets):
- Trypsin - large hydrophobic e.g. Phenylaline
- Negative Coo- in pocket attracts positively charged side chains
- Chymotrypsin - Lysine, arganine
- Large pocket lined with hydrophobic residues
- Elastase - small neutral alanine, serine
- Only small side chains can enter
- Thrombin - arganine, glycine
All are initially inactive
Aspartate, Histidine and Serine (catalytic triad) are positioned to create a cleavage site - forms a nucleophile. Undergoes acid/base catlysis and covalent catalysis
Protease Reaction Mechanism

Related discussions on The Student Room
- Bio a level help - end product inhibition »
- Biology question enzyme affinity »
- Importance of enzymes in biology synoptic essay »
- biology »
- biology aqa alevel amino acid question »
- Exam help gcse bio edexcel »
- Test revision notes gcse edexcel higher biology »
- A level biology AQA »
- 25 mark essay question »
- biology a level electrophoresis question »
Comments
No comments have yet been made