Biological Membranes
0.0 / 5
- Created by: rosieevie
- Created on: 14-01-17 21:01
Importance of Membrane Proteins
- 20% human genome codes for membrane proteins
- Important interface of cell
- Dictate how cell reacts with environment
- Important drug targets
- Faulty membrane proteins cause:
- Cystic fibrosis
- Malignant hypothermia
1 of 14
Eukaryotic Cell Membranes
- Internal organelles bounded by membranes
- Membranes invisible to light microscopes (okay with electron)
- Two major components:
- Lipids -small non-polar soluble molecules = permeability barrier
- Proteins = biological properties e.g. transport, signalling, adhesion
2 of 14
Membrane Lipids - Phospholipids
- Glycerol-based molecules e.g. phosphatidycholine
- Sphinogosine-base molecules e.g. sphingomyelin
- Also contain sterols e.g. cholesterol
3 of 14
Membrane Lipids

- Ampipathic - contain polar and non-polar groups
- In an aq. environment hydrophobic effect occurs:
- Driven by water
- Hydrophobic mol. interfere with water's hydrogen bonds
- Water will squeeze lipids together to minimise interactions of hydrophobic areas to water
- Spontaneous formation of lipid bilayers (act as permeability barriers for polar/large mols) or self-sealing vesicles in water
- Membranes not spontaneously produced - only build from inserting lipids into new bilayers
- Enzymes move phospholipids to either side - flippases (to cytoplasmic side) and floppases (to extracellular fluid) = transverse diffusion
4 of 14
Liquid Crystalline Phase
- Membranes have lots of lipids with acy chains containing cis-double bonds
- Cis-bonds prevent close acyl chain packing = motile
- Liquid crystaline phase = lipids can move around in bilayer (low melting temperatures)
- This is called lateral diffusion - same side
- Essential for function at low temperatures
- Low low temperatures = gel phase and acyl chains frozen - lipids cannot move
- Membrane proteins must undergo conformational changes and interact with adjacent proteins
5 of 14
Membrane Proteins
- Provide control for the internal cell/organelle environment
- Transporters, channels, receptors
- All can be transmembranous
- a-helical conformation (stabilised by H bonds) means they can cross hydrophobic core
- Transmembrane domain residues are hydrophobic
- End residues are polar
- R-groups project outwards = anchorage in bilyaer
- Single helix cannot form route for polar solutes as they are too big/polar
- Amphipathic helicases (~4) cluster w/ polar resiudes on inside = route
6 of 14
Gated Ion Channels
- Makes membrane selective
- Charge at beginning of channel = only opposite charge attracted
- Gate required to stop flow of ions (depending on electrochemical gradient)
- Helicases rotate to expose small polar groups and open channel
- When gate closed hydrophobic groups on inside = no polar molecules
- Ligand binds to cause helicases to rotate
- Cannot
- Move ions/mols against concentration gradient
- Facilitate large molecules (ions would sneak through, disrupting conc. gradients)
7 of 14
Transporters
- 2 gates - neither open at same time = no major change in ion gradients
- Solute recognition site - solute binds causing 1st gate to shut and 2nd to open

8 of 14
Facilitated Diffusion
- Transporter undergoes conformational change e.g. rocking
- Molecule binds tightly in either direction
- No net transport after equilibrium
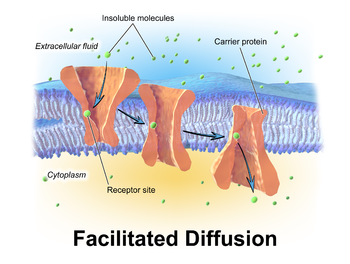
9 of 14
Active Transport
- Against a concentration gradient
- Molcule needs to bind tightly then let go at end
- Requires ATP to overcome attraction force = pull molecule and channel apart
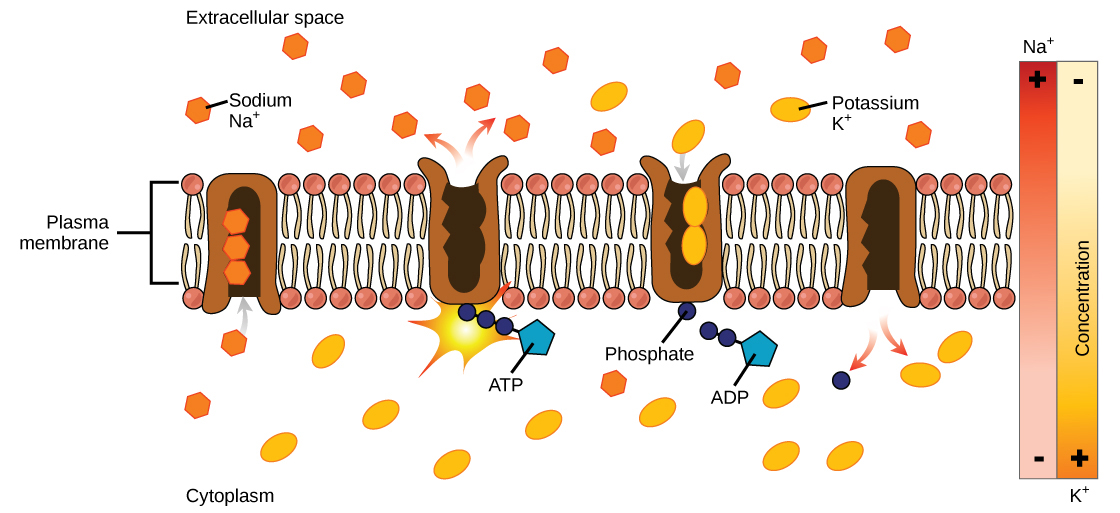
10 of 14
Modelling Membrane Proteins
- Do not readily form 3D structures = few been mapped
- Difficult to disrupt bilayer and isolate proteins
- Most models based on sequence analysis and indirect studies
- You need to form protein crystals and fire x-ray beans to be scattered = diffraction pattern
- Must trap proteins in various conformations = flip-book method
Labelling Studies
- Identify surface exposed proteins using membrane impermeant reagents - covalent modifers modifying specific residues on outside = analyse sequence
- Identify transmembrane reagents using hydrophobic labelling reagents
- Must first break membrane to allow reagent to dissolve in bilayer
- Use detergent = collapse and micelles form
- Proteins react with detergent = protected in shape
- Can be seperated and purified
11 of 14
Modelling Membrane Proteins - Sequence Analysis
- Reveal potential transmembrane helices
- Amphipathic helicases hard - polar amino acids break up line of non-polar ones
- Hydropathy Plots:
- Each amino acid assigned a value for its hydrophobicity (energy required to move residue from aqueous to inside bilayer)
- Higher value = more hydrophobic
- Average length of transmembrane helix 20 residues -> look for 20 consecutive hydrophobic amino acids
- Computer plots = potential transmembrane helicases in peaks
- No idea of orientation of proteins
- Combo of this and labelling studies required
12 of 14
Glycosylation of Membrane Proteins
- Glycophorin A etc. are glycosylated on extra cellular surface
- Precise pattern depends on battery of inherited glycosylation enzymes of an individiual
- Gives rise to ABO blood group system
13 of 14
B-Barrel Structures
- Less common
- Formed of antiparralell B-sheets -> barrel structure
- Often got alternating hydrophobic and philic residues (polar inside and non-polar outside = hydrophilic pore/porin
- Not found in plasma membrane - ion concentration gradient issues
- Found in mitochondria
- Porous membrane preserves intermembrane space = required for mitochondria function
- Found in outer membranes of gram negative bacteria e.g. E.coli
- Live in hostile environmnets with protases
- Protects the inner layer which contains transporters
- Outer layer allows mols. to enter periplamic space down conc gradient
- Prevention of proteases entering
14 of 14
Related discussions on The Student Room
- Hello and i need help with this question »
- a level biology help »
- really stuck on uni essay »
- What are the stalked particles on the inner membrane of the mitochondria ? »
- AQA A level bio help? »
- AQA A-Level Biology Paper 3 [21st June 2023] Exam Chat »
- Is my explanation correct? »
- Edexcel A Level Biology B Paper 2: 9BI0 02 - 17 Jun 2022 [Exam Chat] »
- Biology essay - importance of proteins in living organisms »
- WJEC A-Level Biology Unit 3 (A2) [7th June 2023] Exam Chat »
Similar zoology resources:
0.0 / 5
0.0 / 5
0.0 / 5
0.0 / 5
0.0 / 5
0.0 / 5
0.0 / 5
0.0 / 5
0.0 / 5
Comments
No comments have yet been made