Microbiology
- Created by: cmoore16
- Created on: 01-04-16 17:20
Bacterial cell structure
Bacteria: prokaryotic cells. The cell walls of Bacteria, contain peptidoglycan.
Archaea are prokaryotic cells. The cell walls of Archaea =contain pseudo-peptidoglycan. Archaea often live in extreme environments.
Eukarya have eukaryotic cells. Not all Eukarya possess cell walls. Cell walls from Eukarya that posess cell wall do not contain peptidoglycan.
Prokaryotes: absence of internal compartmentalisation by membranes
Eukaryotes: presence of membrane bound organelles such as the nucleus, mitochondria and chloroplasts
- Bacteria can sense and respond quickly to their environment(despite lack of organelles)
- Most bacteria between: 0.5-1μm x 2-5 μm. Oscillatroia are 8x50μm.
A typical bacterium usually consists of:
- cytoplasmic membrane surrounded by peptidoglycan cell wall
- a fluid cytoplasm that contain the chromosome (nucleoid) and numerous ribosomes
Bacterium internal components
Nucleoid: DNA not surrounded by nuclear membrane (no nucleus). Single circular chromosome. DNA packaged in proteins.
Ribosomes: ~15000 per cell. No separation of function:
- Eukaryotes transcription in nucleus, translation in cytoplasm
- Prokaryotes: transcription and translation take place in the cytoplasm: ribosomesstart translation on RNA as it is being transcribed.
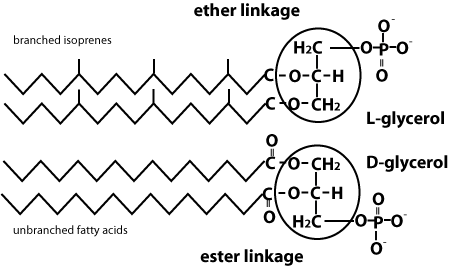
The lipids in Archaean cell membranes are different:
- Ether link
- Branched hydrocarban
- Features that may provide strength in a more extreme environment
Structures that are not universal amongst bacteria
1) Storage compounds: C: poly-hydroxybutyrate, glycogen. P: polyphosphate granules
2) Gas vesicles: bouyancy regulators (Made of proteins gas permeable, but not water permeable controls floating properties).
3) Magnetosomes - MamK (actin homologue) controls the organisation of magnetite particles.
- Magneto-taxis in the northern and southern hemispheres aids cells in efficiently finding their optimum oxygen concentration (O2) at the microaerobic oxic–anoxic transition zone in water columns.
4) Endospores: Some Gram+ bacteria have 2-phase life cycle :
- vegetative cell & and endospore
- sporulation -formation of endospores
- germination- return to vegetative growth
- hardiest of all life forms
- withstand extremes in heat, drying, freezing, radiation & chemicals
- adaptation
Structures that are not universal amongst bacteria
- Dormant cell: 15% water vs 70% in normal cells
- No metabolism
- Extremely resistant to extreme environment (heat, desiccation, radiation, acids, chemical disinfectants)
- Thick coat, DNA, few ribosomes, few enzymes
- May lay dormant for thousands of years, then germinate in thepresence of water and nutrients
- Made in adverse conditions like the lack of nutrients.
External structures
1) Flagellum: bacterial movement; rotation of the flagella results in a propeller motion that moves the bacterial cells forward.
2) Pili (fimbrae):
- more numerous than flagella (10-250 per cell)
- shorter than flagella ~1μm
- function: attachment or sex (occasionally motility)
Structures that are not universal amongst bacteria
4) Axial filaments: Also called Endoflagella
- Periplasmic – between cell wall and outer sheath
- in spirochetes
- Anchored at one end of a cell
- Rotation causes cell to move – like a corkscrew
5) Glycocalyx coat:
- complex carbohydrate
- polysaccharide + protein= glycoprotein coat
- protein coat
- Loosely attached coat: slime - Firmly attached coat: capsule
- Protection against: drying, phagocytosis, buffer between cell andenvironment, Adhesion, Cell-cell communication, Modulation of immune responses, Pathogenicity.
Biofilms
In the environment most bacterial population are part of biofilms.
How is the biofilm built?: require: surface ( solid-liquid or gas-liquid interface), moisture & nutrients
Primary colonisers form first and either divide or form microcolonies. Secondary colonizers coadhese.
Biofilm properties
Biofilm communities can be formed by a single bacterial species, but innature biofilms almost always consist of rich mixtures of many species of bacteria, as well as fungi, algae, yeast and protozoa. Biofilm formation in sea:
- Bacerial cells settle onto hard surface.
- Cells proliferate and secrete slime, forming a biofilm.
- Some cells detach.
- Protists graze on bacteria.
- If enough biofilm forms, large organisms can attach to it.
Biofilms are held together by EPS : Extracellular Polymeric Substance, which often contains extracellular polysaccharides (slime).
Some of the cells within the biofilm produce EPS that will act as a “glue” to generate complex, 3D structures.
This slime layer of EPS and bacteria entraps particulate materials such as clay, organic materials, dead cells and precipitated minerals, adding to the bulk and diversity of the biofilm habitat.
Characterisation of biofilms
In the biofilms cells can coordinate their behaviour by intercellular communication via signalling molecules.
Biofilms are less susceptible to antibiotics - Even when biofilms are treated for prolonged periods of time or with elevated antimicrobial concentrations, a small fraction of the population persists.
Beneficial biofilms: Bioremediation : waste water treatment plants and healthy gut flora.
Biofilms and Infections:
- Mixed biofilms on the artificial surface of catheters or other implants.
- Biofilms play a role in Bacterial Endocarditis (infection of the inner heart and its valves).
- Biofilms form frequently in patients with Cystic Fibrosis.
- Biofilms also play a role in Legionnaire's disease.
- Due to the morphology of biofilms, bacteria capable of forming them are highly resistant to antibiotics, making treatment very difficult.
Bacterial communication in biofilms
Quorum Sensing: Regulation of gene expression in response to fluctuations in population density. Single bacterium <--> A large group of bacteria ( Quorum).
- V. fischeri (Gram-negative bacterium) lives in the light-sensing organs of the bobtail squid for camouflage.
Two key components: I proteins & R proteins:
- LuxI: produces the signal molecule (often called autoinducer, AI).
- LuxR: transcriptional activator
- Upon binding AI, LuxR activates a set of genes including the lux genes for bioluminescence.
How Pathogens Use Quorum Sensing? The bacteria appear relatively innocuous as they quietly grow in number.
- These changes culminate in an infection that can ambush and overwhelm our immune system defenses.
- When their population reaches a certain
- level, instant changes occur in their: Behavior, Appearance, Metabolism
Bacterial cell signalling
Two component signal transduction system is important for signalling in Gram-positive bacteria. Oligopeptide mediated quorum sensing in Gram-positive bacteria:
Bacterial two component system: H: His of a sensor kinase and D: Aspartate of the response regulator
Comparison of signalling in Gram-positive and Gram
Bacterial shapes
Average size: 0.2 -1.0 μm Å~ 2 - 8 μm.
Basic shapes: Spherical, rod, spiral
Unusual shapes: Star-shaped Stella, Square Haloarcula
Most bacteria are monomorphic, a few are pleomorphic.
Pairs: diplococci, diplobacilli
Clusters: staphylococci
Chains: streptococci, streptobacilli
Shape is genetically determined
- cell wall (exocytoskeleton)
- ***** the cell wallà spheroplast/ protoplast
- genes involved in cell wall synthesis
Bacterial cell wall - Gram positive and negative
Peptidoglycan:
- Confers rigidity to bacterial cell walls.
- Backbone of alternating sugars: N-acetylglucosamine, N=acetylmuramic acid (unique)
- Amino acid side chains on NAM: D amino acids, Diaminopimelic acid
Gram-positive cell wall:
- Thick peptidoglycan (60-90% of wall)
- Surface studded with proteins
- No lipids present in the cell wall
- Molecules pass freely through peptidoglycan so it does not trap molecules unless they are tethered to cell membrane or wall
Gram-negative cell wall:
- 2 layer wall: thin layer of peptidoglycan (10-20%) & outer membrane/permeability barrier
- Periplasm: gelatinous material between the outer membrane, peptidoglycan and cytoplasmic membrane. Contains proteins needed for transport of material to and from cells.
Bacterial cell wall 2
Outer membrane:
- Phospholipid bilayer
- Lipopolysaccharide: lipidA portion is embedded in outer membrane, polysaccharide - Only in gram negative bacteria
- extends outward from surface
- Surface of membrane is studded with proteins
- The membrane is semi-permeable containing protein channels like porins.
Some bacterial groups lack typical cell wall structure i.e. Mycobacterium and Nocardia.
Some have no cell wall i.e. Mycoplasma - cell wall is stabilized by sterols
Cell walls from Archaea lack peptidoglycan:
- N-acetyl talosaminuronic acid
- Archaea have no D-amino acids in their cell wall
Bacterial cell wall growth
How does the cell wall grow?
- NAM and NAG are synthesized in the cytoplasm
- attached to a membrane carrier molecule called bactoprenol transported across the cell membrane
- Links in the peptidoglycan have to be broken
- Autolysins: break glycosidic bonds and peptide cross-bridges
- New peptidoglycan monomers (NAM-NAG dimer) inserted
- Transglycosidase enzymes
- Peptide crosslinks resealed
- Transpeptidase enzymes: penicillin binding proteins (PBPs) (inhibited by penicillins)
Bacterial cytoskeleton 1
Eukaryotic cells have four main types of cytoskeletal elements:
- microtubules, composed of tubulin
- microfilaments, composed of actin
- intermediate filaments
- molecular motors
Bacterial cytoskeletal elements:
- tubulin homologue: FtsZ
- actin homologue: MreB, Mbl proteins; ParM, MamK
- intermediate filaments: CreS
- molecular motors – not found in bacteria
Cell shape in bacteria is controlled by: 1) The Actin homologue, MreB
- MreB polymerises into filaments in the presence of ATP or GTP
- MreB direct the cell shape by localizing the proteins in a “peptidoglycan factory”
Bacterial cytoskeleton 2
Correlation between nonspherical shape and the presence of at least one MreB homologue.
Location of cell wall growth:
- Spherical bacteria: Cell wall extension only occurs at the middle of the dividing cell using FtsZ as a scaffold
- Rod shaped: Part of the cell wall synthesis takes place throughout the entire cell length (alongside MreB cables).
2) Tubulin homologue – FtsZ controls cell division in bacteria by dividing cell.
- FtsZ polymerises into a ring structure in the presence of GTP (GTPase activity)
- FtsZ recruits other proteins, some of which are involved in new cell wall synthesis at the division site.
In addition to the lateral wall extension directed by MreB, cell wall extension also occurs at the middle of the dividing cells using FtsZ as a scaffold.
3) Intermediate filaments in bacteria
Antibiotics - definitions and structural classes
Antibiotic: Chemical produced by a microorganism that kills or inhibits the growth of another microorganism
Antimicrobial agent: Chemical that kills or inhibits the growth of microorganisms
Bactericidal : kills bacteria
Bacteriostatic: inhibit growth and reproduction of bacteria , but does not kill
1) Structural classes of antibiotics:
- Glycopeptide antibiotics - Glycosylated cyclic or polycyclic nonribosomal peptides.
- Tetracyclin Antibiotics - Contain 4 hydrocarbon rings
- Beta-lactam antiobiotics - contains a β-lactam ring in their molecular structures. Includes penicillin derivatives
- Aminoglycoside Antibiotics - inhibit bacterial protein synthesis.
- Macrolide Antibiotics - inhibit the growth of bacteria and are often prescribed to treat rather common bacterial infections.
Antibiotic targets/mechanisms of action
2) Antibiotic targets/mechanisms of action
Antibiotic Resistance mechanisms
3) Antibiotic Resistance mechanisms
(a) impermeable barrier blocks antibiotics (blue spheres) because the bacterial cell
envelope is now impermeable to the drug.
(b) Target modification alters the proteins inhibited by the antibiotic, so the drug cannot bind properly.
(c) Antibiotic modification produces an enzyme that inactivates the antibiotic.
(d) Efflux employs genes coding for enzymes that actively pump the antibiotic out of the cell.
Penicillin
Acquiring Antibiotic Resistance
Antibiotic resistance can be acquired in two basic ways. In vertical transmission, a bacterium accumulates errors or mutations in its genome during replication; some of those changes (red) give the ability to resist antibiotics and are passed on to subsequent generations. In horizontal transmission, resistant genes are swapped from one microbe to another.
Bacterial motility
Swarming is the multicellular movement of bacteria across a surface and is powered by rotating helical flagella.
Swimming is the movement of individual bacteria in liquid, also powered by rotating flagella.
Twitching is surface movement of bacteria that is powered by the extension of pili, which then attach to the surface and subsequently retract, pulling the cell closer to the site of attachment.
Gliding is active surface movement that does not require flagella or pili and involves focaladhesion complexes. Process:
- Large focal adhesion complexes extend from the cell and connect the extracellular surface to actin-like cytoskeletal filaments.
- Proteins attached to the intracellular portion of the focal adhesion push backwards and move the focal adhesion along the cytoskeletal filament to move the cell forward – whilst they stay fixed at fixed position with respect to the substratum.
Sliding is passive surface translocation that is powered by growth and facilitated by a surfactant.
Bacterial motility 2
Bacterial swimming requires flagella. Flagellum components:
- Basal body attached to cell membrane and wall.
- A hook anchored in the basal body.
- Long fine filament, a helical protein made of subfilaments (flagellin).
Rotation of the basal body and flagellum: Energy required comes from proton motive force:
- H+ movement across membrane through
- Mot complex drives rotation of flagellum
- 1000 H+ must be translocated per single rotation of flagellum
Bacterial chemotaxis = ability of a cell to sense external concentration of a chemical species and migrate (directed movement) towards/away from higher concentrations
Phototaxis: Movement toward light. Phototrophic organism orients itself most sufficient for photosynthesis.
Switch between CCW and CW rotation of the flagella : FliM switch protein
Diversity of microbes 1
Photosynthetic: photoautotrophs, which means that they are able to synthesize food directly from carbon dioxide and water using energy from light., 99% of life
Chemosynthetic: biological conversion of one or more carbon molecules (usually carbon dioxide or methane) and nutrients into organic matter using the oxidation of inorganic (e.g. hydrogen gas, hydrogen sulfide) or methane as a source of energy.
Classification of Microorganisms: (Name - Energy source - carbon source)
- photoheterotroph - light - organic compounds
- photoautotroph - light - carbon dioxide
- Chemoorganoheterotroph - organic compounds - organic compounds
- chemolithotroph - inorganic compounds - organic compounds
- chemolithoautotroph - inorganic compounds - carbon dioxide
Diversity of microbes 2
What is required for life?
- Essential elements: C, O, N, S, P, trace elements like Fe
- Essential molecules: H2O
Energy sources to survive and grow.
- Phototrophic organisms - Sunlight generates energy.
- Chemotrophic organisms - Breakdown of molecules generates energy.
Higher organisms need lots of energy
- Obtained from O2 and high energy carbon sources
- Plants use high energy wavelengths of light.
Bacteria are more versatile
- Various molecules can be used instead of O2 and carbon sources
- Photosynthetic bacteria can use different energy wavelengths.
Generation and Consumption of Energy in the Cell
Chemical energy is obtained from redox reactions
- ΔG = - nF ΔE
- F = Faraday constant (96.4 J mV-1 mol-1)
Energy conservation in the microbe
Microbes must obtain energy to survive and grow
Energy must come from environment (e.g. Light or available molecules)
During the production of energy the microbe MUST keep cellular components balanced.
- NAD+/NADH equilibrium
- Quinol/Quinone pool equilibrium
- Proton motive force equilbrium
Energy conservation in the microbe
Microbes must obtain energy to survive and grow
Energy must come from environment (e.g. Light or available molecules)
During the production of energy the microbe MUST keep cellular components balanced.
- NAD+/NADH equilibrium
- Quinol/Quinone pool equilibrium
- Proton motive force equilbrium
Microbe central Metabolic Pathways
How do organisms obtain energy? - Breakdown of chemical compounds
- Chemotrophy
- Respiration
- Fermentation
- Harvesting of external energy - Phototrophy
- Photosynthesis
- Photophosphorylation
ATP Generation in Fermentation
Fermentation:
- A single substrate is sequentially oxidised and then reduced.
- As part of the process electron shuttles (typically NADH) are reduced and then oxidised.
- Fermentation does not yield as much energy as respiration.
- Historically described as metabolism in the absence of oxygen, sometimes used simply to describe microbiological growth.
- Important biotechnological process.
Fermentation is an ancient mode of metabolism, and must have evolved with the appearance of organic material on the planet. (predates photosynthesis)
No outside electron acceptors are involved; no membrane or electron transport system is required; No ATP synthase needed
Fermentation is metabolism where energy is derived from the partial oxidation of an organic compound using organic intermediates as electron donors and electron acceptors.
All ATP is produced by substrate level phosphorylation.
Glycolysis as a source of, pyruvate ATP and NADH
Input: 2 ATP, 2 NAD+
Output: 4 ATP, 2 NADH
Net: 2 ATP & 2 NADH
Note: Fermentation has to be happen before the cell starves itself. Molecule of glucose phosphorylated twice by 2 p's.
Lactic Acid Bacteria (e.g. Streptococcus, Lactobac
Obligately fermentative, aerotolerant, Gram-positive bacteria that:
- ferment sugars with lactic acid as the major product
- Lactobacilli are relatively acid-tolerant growing well down to pH4.
- Lactobacilli are not pathogenic (their acid production kills other organisms).
- Acid production by Streptococci is a dominant factor in tooth decay.
- Only lactobacilli and some enteric bacteria have the capacity to metabolise the disaccharide lactose (glucose + galactose).
The lactate dehydrogenase reaction disposes of reductant but does not itself generate ATP. The lactate dehydrogenase reaction must therefore be linked to glycolytic ATP production.
Fermentations Involving Pyruvate - Ethanol Ferment
Yeast are facultive anaerobes. The yeasts used in the production of alcoholic beverages and industrial ethanol are strains of the bakers’ yeast Saccharomyces cerevisiae. In breadmaking the CO2 produced by fermentation is used to raise the dough.

Production of alcohol kills off competing strains who aren't alcohol tolerant.
Respiration
During respiration Pyruvate is converted to acetyl-CoA. Acetyl-CoA feeds into theTCA cycle.
Catabolic reactions producing ATP in which either organic or inorganic compounds serve as electron donors and organic or inorganic compounds serve as electron acceptors. The reactions of respiration are associated with the cytoplasmic membrane, and ATP is generated by oxidative phosphorylation.
Aerobic respiration in E. coli
E. coli faces a number of challenges as a gut bacterium. Respiration must be regulated to meet cell demand.
Log phase, High aeration ATP required, maximise P/O ratio. Cytochrome bo oxidase expressed.
Stationary phase, low aeration ATP less important, lower P/O ratio. Cytochrome bd oxidase expressed.
E. coli contains 3 quinol oxidases (cytochrome bo3, bd1and bd2).
- bo oxidase has a lower affinity for oxygen and a proton pumping mechanism
- bd oxidase has a higher affinity for oxygen and no proton pumping mechanism
Bacteria use denitrification for respiration and protection during infection.
Obtaining energy
CHEMOTROPHIC
Respiration - Molecule (1) oxidised, Molecule (2) reduced
- + can gain large amounts of energy
- - Requires 2 substrates
Fermentation - Substrate (1) oxidised, breakdown product of substrate(1) reduced.
- + Only one substrate required
- - low energy yield
PHOTOTROPHIC
Non-Photosynthetic - Light is used to generate chemical energy (ATP)
Photosynthetic - Light is used to generate energy (ATP) and also oxidise substrates for reductive energy (NAD(P)H)
Phototrophy
Conversion of light energy into chemical energy in the form of ATP
- Nonphotosynthetic photophosphorylation - Archea e.g. Halobacterium salinarum
- Anoxygenic photosynthesis - Purple non-sulfur bacteria e.g Rhodobacter
- Purple/Green Sulfur bacteria e.g Chromatiaceae
- Oxygenic photosynthesis - Cyanobacteria e.g Nostoc, Oscillatoria
Halobacterium salinarum - A model organism for the halophilic branch of the archaea.
- Lives in highly saline environments
- Mass cultures recognized by their typical colour, which originates from bacterioruberin.
- Best studied ion pump is Bacterhodopsin
Bacterhodopsin:
- Major photosynthetic protein of Halobacterium salinarum.
- Converts the energy of "green" light into an electrochemical proton gradient, Which in turn is used for ATP production by ATP synthase.
- A light-driven proton pump, transporting protons out of the cell - vectorial catalysis
Photosynthetic Bacteria
Nonphotosynthetic photophosphorylation in archae
- Outer membrane proton pump
- ATP generating
- No reducing equivalents – no CO2 fixation
- Not photosynthetic, but these pumps allow the bacteria to survive in conditions of low oxygen – requires oxygen for retinal synthesis
Different bacteria utilise different wavelengths of light (Use Winogradsky Columns):
- The absorbance spectra of different bacteria are tuned to capture specific wavelengths of light
- Cyanobacteria and algae all utilise between 400-700 nm, with others harvesting around these wavelengths.
Chlorophylls Absorb Light Energy for Photosynthesis
- Most important light absorbing pigments in the thylakoid membranes are the chlorophylls
- Resemble protoporphyrin of hemoglobin but coordinate Mg2+ not Fe2+
- All chlorophylls have a long phytol side-chain
Light Harvesting Complexes of Rhodobacter
- There are two light-harvesting complexes in rhodobacter membranes
- These can be separated by density gradient centrifugation
- Light Harvesting complex LH2 contains carotenoids
- Light Harvesting complex LH1 contains carotenoids and the reaction centre, which contains chlorophyll and quinols.
Light harvesting complexes collect energy from light and transfer it to the reaction centre.
Proton pumps and electron flows
Protons are Pumped by the Cytochrome bc1 Complex
This is the same cytochrome bc1 complex used in respiration but since there is no oxygen cytochrome c moves back to the RC and not to terminal electron acceptors.
Cyclical Electron Flow in a Reaction Centre
The need for high energy electrons to feed into quinol pool means that these bacteria are not truly phototrophic.
Photosynthesis
Photosynthesis is the conversion of light energy into chemical energy that can be used in the formation of cellular material from CO2.
Catabolism = Light reaction (Generation of ATP/NADH)
Anabolism = Dark reaction (Carbon fixation, uses ATP and NAD(P)H)
CO2 + H+ + e- >>energy>> COOH
Photosynthesis 2
Using H2S as an Electron Source
- The generation of proton motive force is the same cycle as before.
- Electrons can leave reaction centre either through quinol (for proton gradient) or through a ferrodoxin that reduces an FDNAD reductase.
- Electrons from H2S or Fe2+ are used to reduce the Reaction centre.
- The electrons used to reduce ferrodoxin are liberated
Oxygenic Photosynthesis in Cyanobacteria
- Chloroplasts of higher plants evolved from Cyanobacteria
- H2O rather than H2S is the electron donor
- The b6f complex is closely related to the bc1 complex and performs a similar function
Autotrophic CO2 Fixation
In Cyanobacteria and Purple Sulfur Bacteria CO2 is assimilated via a cyclical pathway known as the Calvin Cycle.
- Each molecule of CO2 fixed costs 2NADPH and 3 ATP
Photosynthesis 3
Chlorophylls Absorb Light Energy for Photosynthesis
- Most important light absorbing pigments in the thylakoid membranes are the chlorophylls
- Resemble protoporphyrin of hemoglobin but coordinate Mg2+ not Fe2+
- All chlorophylls have a long phytol side-chain
Purple Non-sulfur Bacteria - Phototropic but not photosynthetic
- Phototrophic members of the gamma division of the Proteobacteria
- Examples include Rhodobacter capsulatus and Rh. Sphaeroides
- Metabolically versatile – can fix N2 and CO2
- Another close relative of eukaryotic mitochondrial ancestor (bc1 complex)
- Intensely studied bioenergetic mechanisms
Oxygenic Photosynthesis in Cyanobacteria
- Chloroplasts of higher plants evolved from Cyanobacteria
- H2O rather than H2S is the electron donor
- The b6f complex is closely related to the bc1 complex and performs a similar function
Related discussions on The Student Room
- Eligible for Microbiology Degree? »
- what are the career prospects for the following degree »
- No idea where to take my life after University »
- can I goto microbiology after having degree in master of zoology »
- Help me choose, dentistry or biology? »
- Urgent please help »
- Immunology at Imperial College London »
- Any ideas for what I should do for microbiology placement research project? »
- Is there any university where I can study Mycology? »
- Astrobiology »
Comments
No comments have yet been made