Dna repliction
- Created by: Former Member
- Created on: 16-03-15 14:44
The genetic code
The information carried in DNA is determined primarily by the sequence of nucleotide bases (A,T,G,C)
Ever 3 bases on DNA codes for one amino acid in a protein/polypeptide and the order of these bases determines the protein that is made.
A gene is a length of DNA that codes for a protein/polypeptide.
Humans pocess 30'000 genes, which represents only 10% of the total DNA. Between the genes are non coding regions of DNA, the function unkown.
The functions of are non coding regions while not understood are highly variable and thus are useful for investigations.
DNA tecnology
Amplifying DNA (making large quantities avaiable) - polymerse chain reaction
Comparing DNA- use of specific marker sites using special enzymes (restriction endonucleases) and DNA probes.
Potential problems with PCR: contamination of sample of DNA for amplification
Polymerase chain reaction
The PCR can produce many copies of the selected section of DNA in a very short time.
The PCR mimics the natural process of DNA replication.
The process requires:
- a DNA sample that includes the selected region for replication
- Primers
- A thermostable DNA polymerase enzyme
- Nucleotides
Uses of PCR
- For research like 'the human genome project'
- inforensic sciene- fingerprinting
- for medical research
PCR METHOD
- The DNA section to be amplified is heated to 95 ℃. This breaks the hydorgen bonds and seperates the 2 strands.
- The mixture is cooled to 50-55 ℃ , which allows the primers to bind (anneal) to the start of each strand of the selected DNA region.
- Mixture heated to 73 ℃ (optimun temp for enzyme) DNA polymerase enzyme copies each strand starting at the primers. (the DNA polymerase reads the template strand and matches it with complementary nucleotides)
- Whole cycle repeated - with each cylce the no. of DNA molecules doubles.
PCR..
Taq polymerase is the enzyme commonly used. This enzyme is ideal as it is 'thermostable' - it won't denature at temperatures up to 80 ℃. Thus the enzyme remains stable during high temperatures used to break hydrogen bonds.
How can temp denature enzymes?
heat disrupts and breaks ionic, hydorgen and disulphide bonds that hold the enzyme in shape, disrupting the enzymes active site.
PRIMERS
Primers are short pieces of DNA containing a known sequence of nucleotides which are complementary to the sequence at the start of each region to be copied.
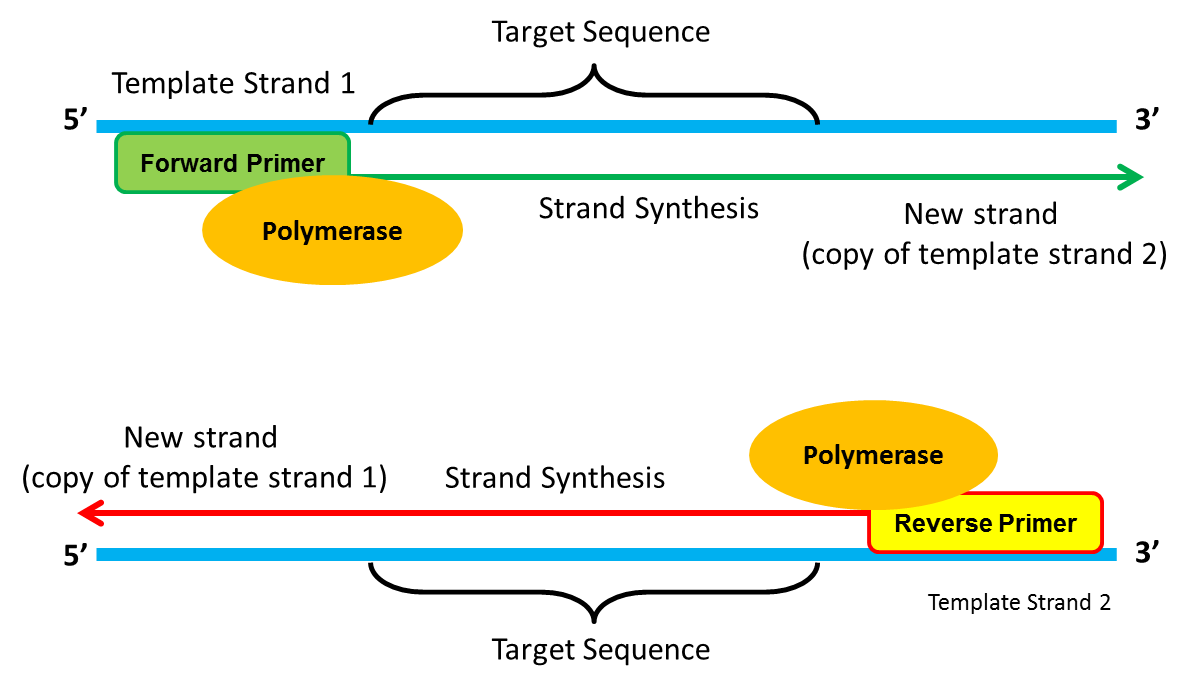
Role of primers:
- stops DNA strands from rejoining
- intiates the action of DNA polymerase
Genetic Markers n restriction enzymes
'A marker' - is a nucleotide sequence that is variable within a population and therfore can be used to determine differnces between indiviuals.
Variable regions of DNA can be recognised:
- Restriction fragment length polymorphisms
- Microsatellite repeat sequences
- Single nucleotide polymorphisms
Restriction endonuclease enzymes- cut DNA at specific base squences. Each enzyme cuts DNA at a specific base sequence- the recognition site. The enzyme cuts the DNA producing a no. of different fragment lengths depending on the number of recognition sites in the DNA. (if 4 sites- 5 fragment lengths produced)
Restriction fragment length polymorphisms (RFLP's)
The different array of fragment lengths produced when a specific restriction endonuclease enzyme is used to cut different DNA molecules.
The variation is due to differences in the no. of recognition sites between indivuals
Each enzyme cuts DNA at a specific base sequence eg:
.
Microsatellite repeat sequences (MRSs)
This marker is composed of a sequence between 1 and 5 bases in length, which is repeated many times (in a length of DNA.)
- Found in non coding regions of DNA.
- The number of times they repeat varies betwen indiviuals so they give unique patterns.
- used in DNA fingerprinting
eg:
DJNIAJATATATATATATIEFBAIFBIA 2 bases AT are repeated over n over
The no. of times these sequences are repeated varies between individuals
Single nucleotide polymorphism (SNP's)
Involves differences in single nucleotides among samples of DNA molecules. The difference between one DNA sample and another is (usually) one nucleotide base (A,T,G or C).
eg: amy has sequence CTTGA while scott has sequence CCTGA
SNP's is used to TEST 4 GENETIC DISEASES as one nucletide change gives rise to existance of different alles eg.csytic fibrosis or sickle cell anaemia
DNA Probes
DNA probes are used to locate a particular section of DNA.
- They can identify whole genes or short sections of DNA
- They have many applications: they can be used to identify the presence of some inherited diseases. (many of wihch are caused by the presence of single alleles)
A probe consists of a short length of single starnded DNA with a specific and known nucleotide base sequence.
The probe will bind by base pairing with any complementary of single stranded target DNA.
A different probe will be used for each marker being looked for.
To allow the postion of target DNA and probe to be detected, the probe is either:
radioactive or fluorescently labelled
DNA PROFILING (fingerprinting)
The DNA of genes that code for the proteins of an indiviual does not show much variation. However the non coding 'junk' DNA that exists between genes is highly variabe. Within these non-coding regions MRS's are found.
Role of fingerprinting in society:
- to solve paternity and relationship disputes
- evolutionary research
- in criminal justice systems
Key features of genetic fingerprinting involve:
- Extracting DNA from a sample. Often PCR is used to amplify DNA for anaylisis.
- restriction enzymes cut the DNA into different sized fragments
- the fragments of DNA are seperated on the basis of size by gel electrophoresis DNA having a negative charge moves towards the positive electrode. Smaller fragments travel further than larger fragments
- DNA treated to make single strand
- The DNA fragments are transfered to a nylon membrane
- Radioactive (or fluorescent) DNA probes are added Only those fragments that have bound to the labelled probe show up.
- Membrane placed on X-ray film, labelled DNA appears as a pattern of bars 'DNA fingerprint' produced
PRINCIPLE OF GENETIC FINGERPRINTING
- Everyone has a unique DNA finger print ( except identical twins)
- the closer the relationship between individuals (or species) the closer the match
- The number of repeats varies between indiviuals
- the greater the no. of repeat sequences the longer the piece of DNA (ATGGGGC/ATGGGGC)
- The smaller fragment size the further it travels in the gel
- The thickness of the bars is an indication of the number of DNA fragments of a particular size. not linked to fragment size
Related discussions on The Student Room
- Biology Paper 2 AQA Triple Higher 2023 »
- GCSE English Language Speech Opinions »
- HELPPPP!!! DNA replication & transcription »
- 25 marker essay biology »
- What is DNA repair »
- Biology OCR past paper question help. »
- PCR A level biology »
- A question about DNA electrophoresis »
- biology a level electrophoresis question »
- DNA mcq »
Comments
No comments have yet been made